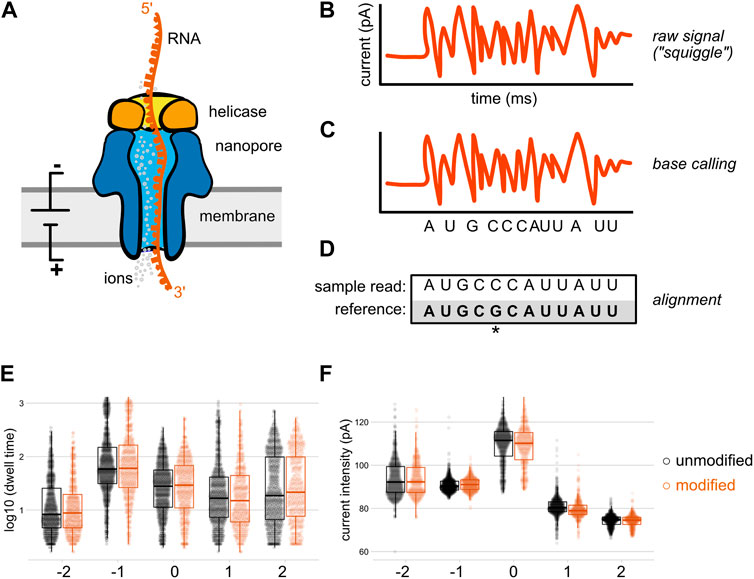
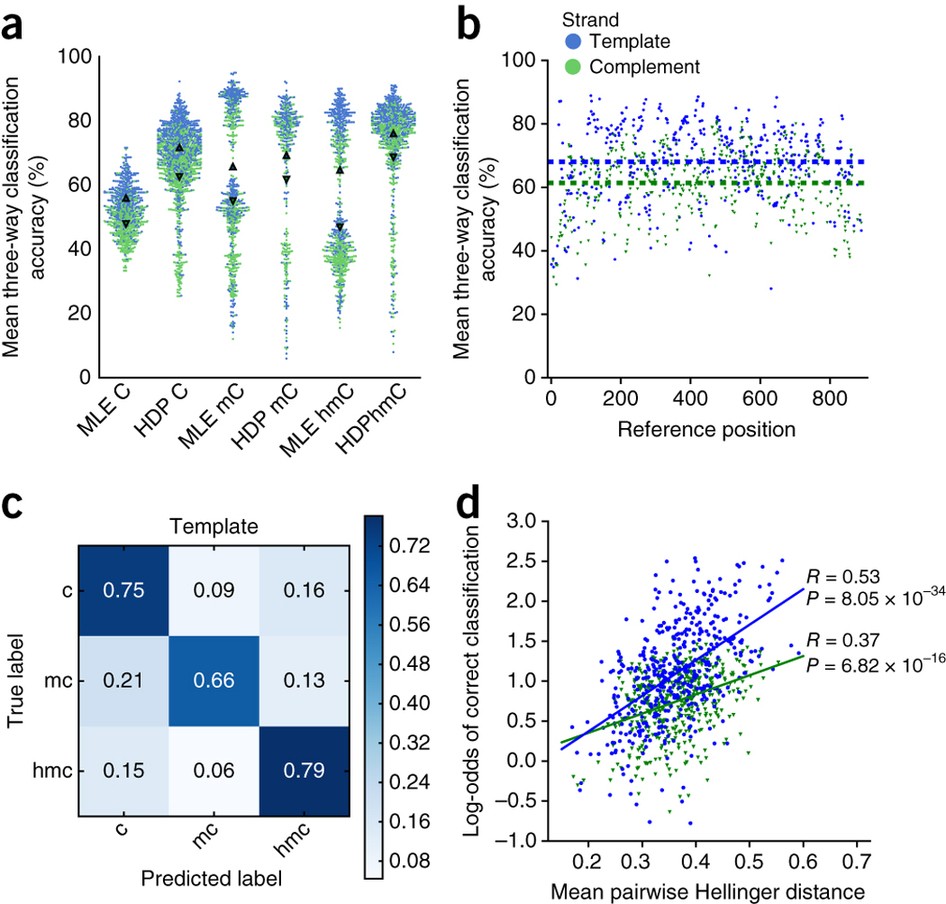
Methylation calling from nanopore data and comparison to gold standard... | Download Scientific Diagram
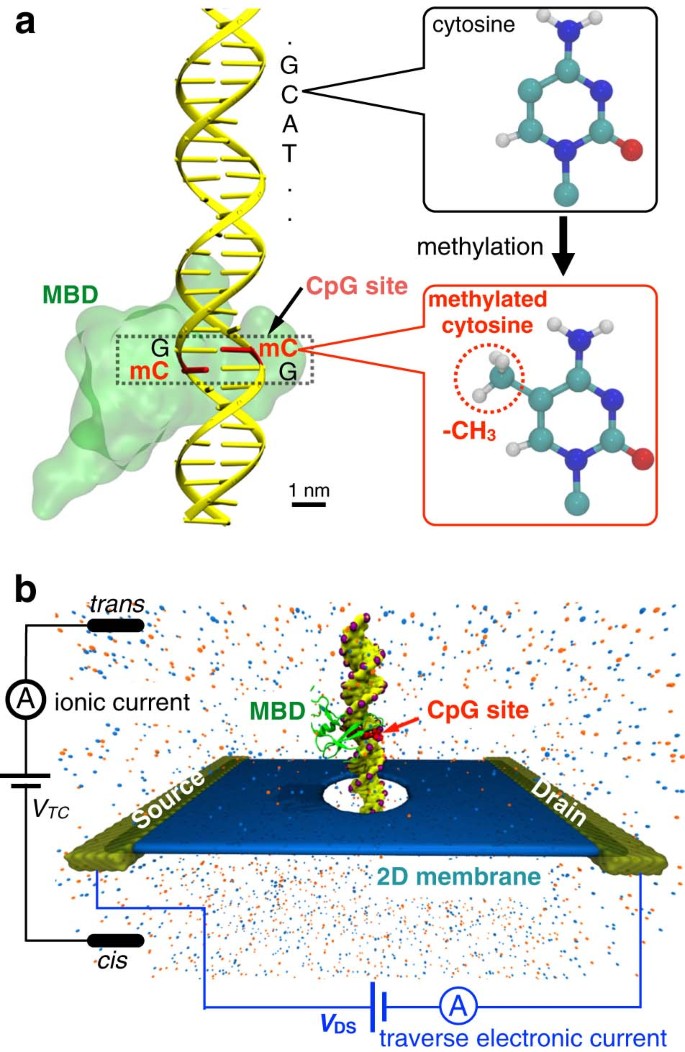
Detection and mapping of DNA methylation with 2D material nanopores | npj 2D Materials and Applications

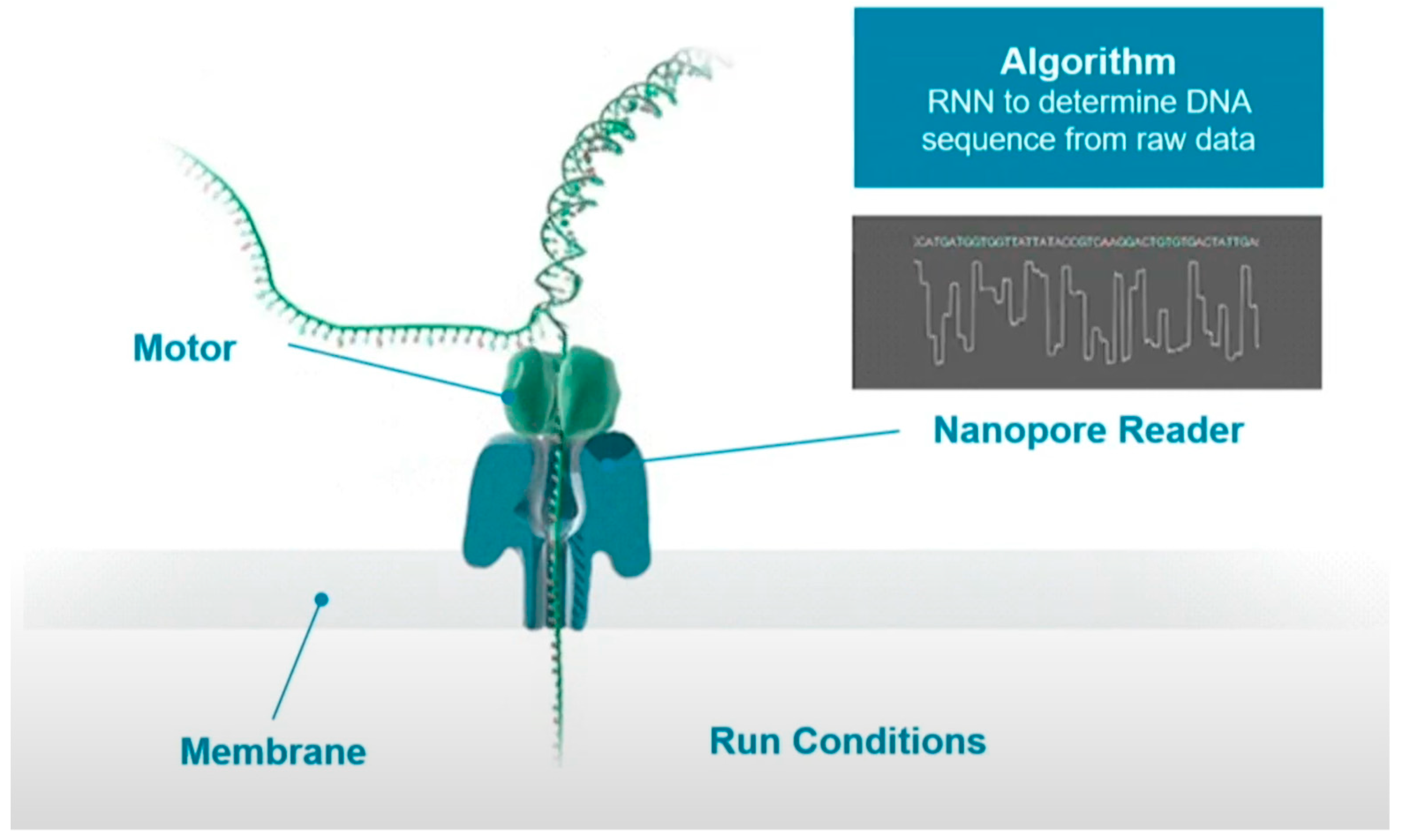
Oxford Nanopore technology update: CTO Clive G Brown unveils latest sequencing chemistry with highest performance to date, Short Fragment Mode and latest methylation performance evaluations

Discovering and exploiting multiple types of DNA methylation from individual bacteria and microbiome using nanopore sequencing | bioRxiv
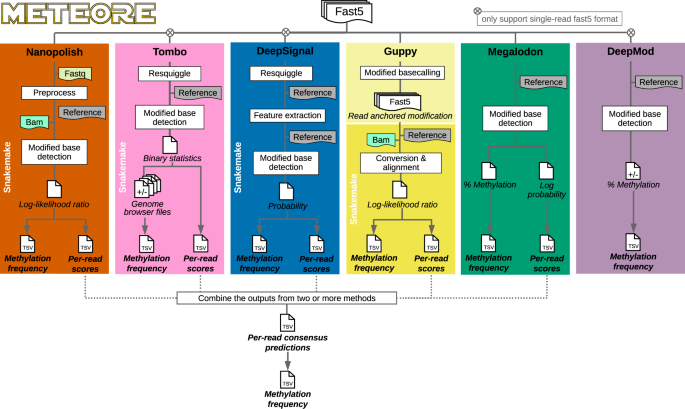
Systematic benchmarking of tools for CpG methylation detection from nanopore sequencing | Nature Communications
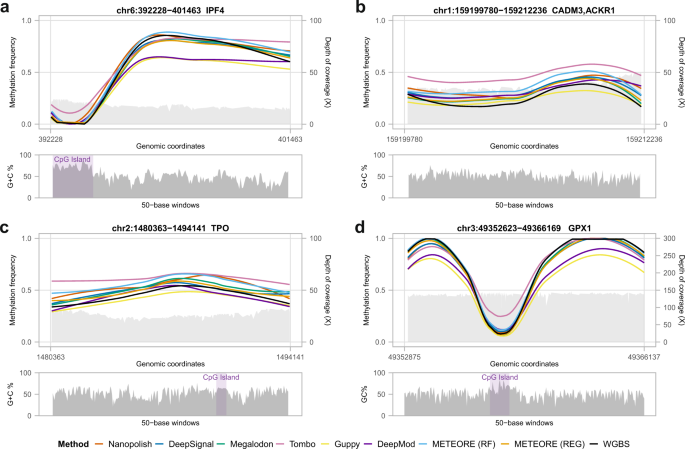
Systematic benchmarking of tools for CpG methylation detection from nanopore sequencing | Nature Communications
Methylation detection with nanopore sequencing: Reduced-Representation Methylation Sequencing (RRMS)
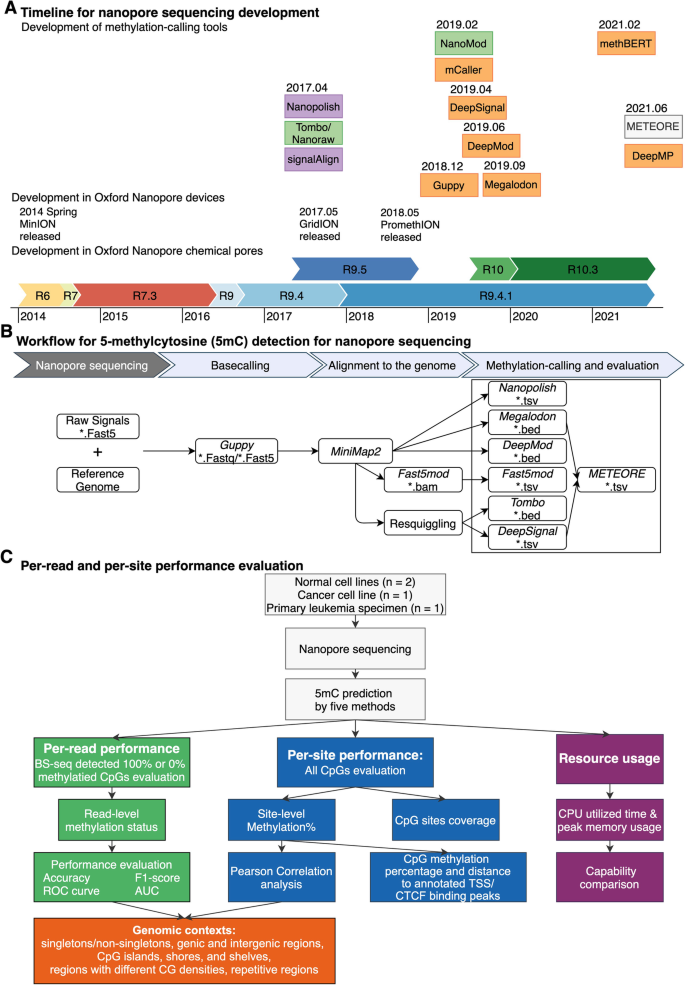
DNA methylation-calling tools for Oxford Nanopore sequencing: a survey and human epigenome-wide evaluation | Genome Biology | Full Text
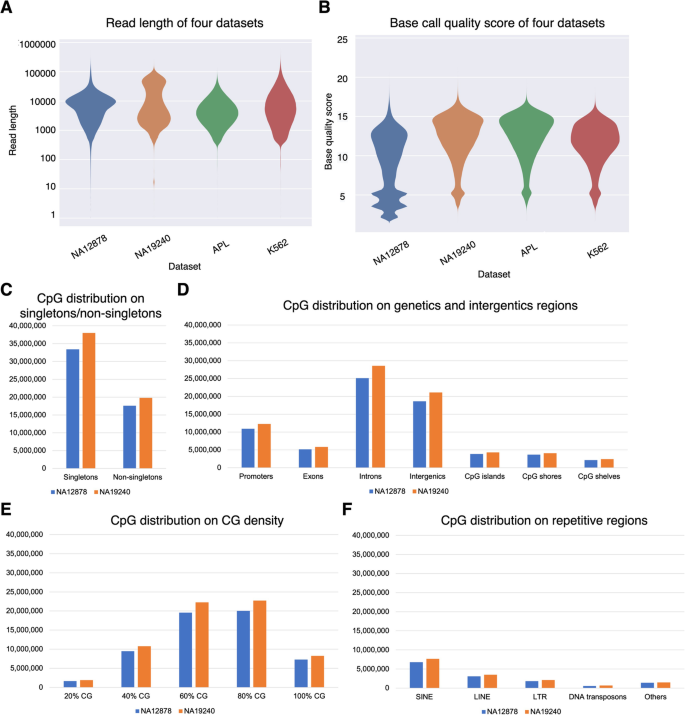
DNA methylation-calling tools for Oxford Nanopore sequencing: a survey and human epigenome-wide evaluation | Genome Biology | Full Text

Discovering multiple types of DNA methylation from bacteria and microbiome using nanopore sequencing | Nature Methods
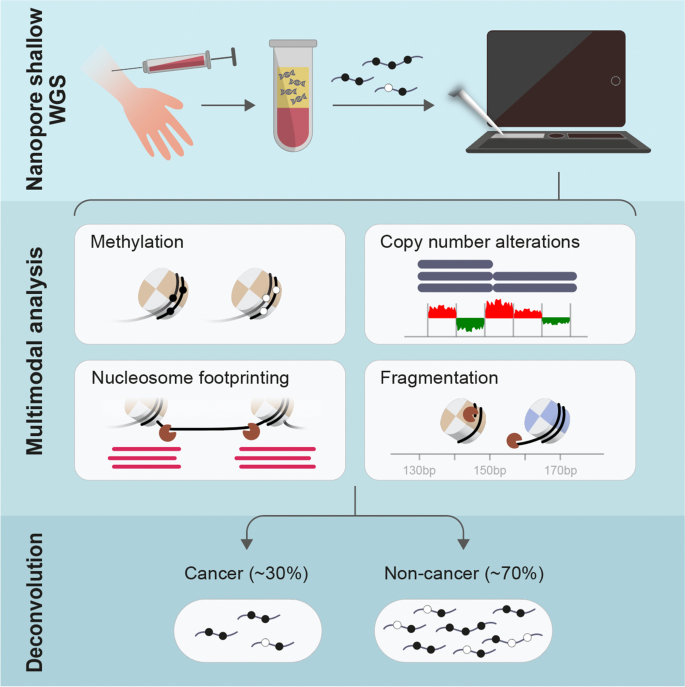
Detecting cell-of-origin and cancer-specific methylation features of cell-free DNA from Nanopore sequencing | Genome Biology | Full Text
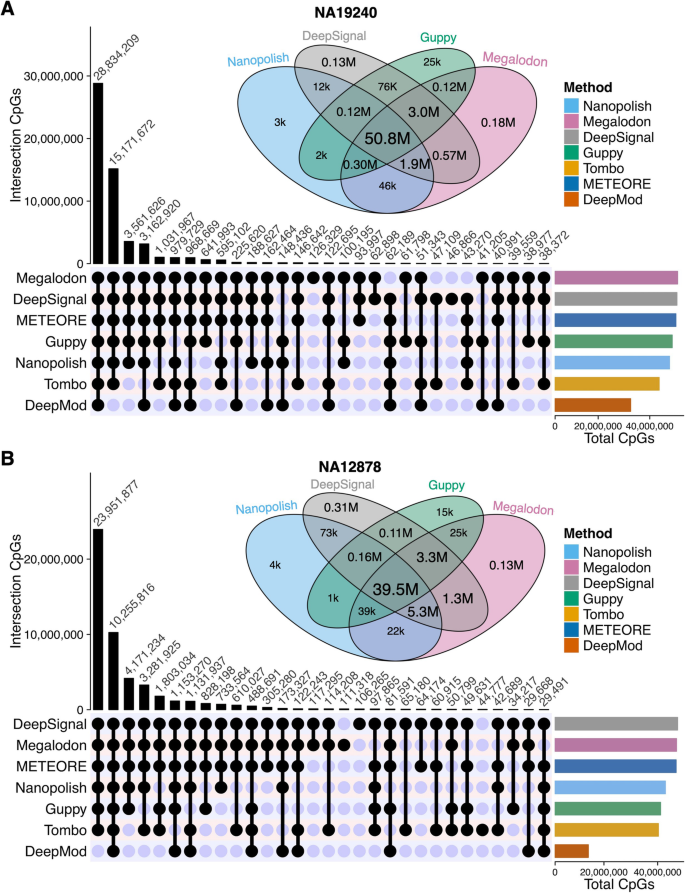
DNA methylation-calling tools for Oxford Nanopore sequencing: a survey and human epigenome-wide evaluation | Genome Biology | Full Text
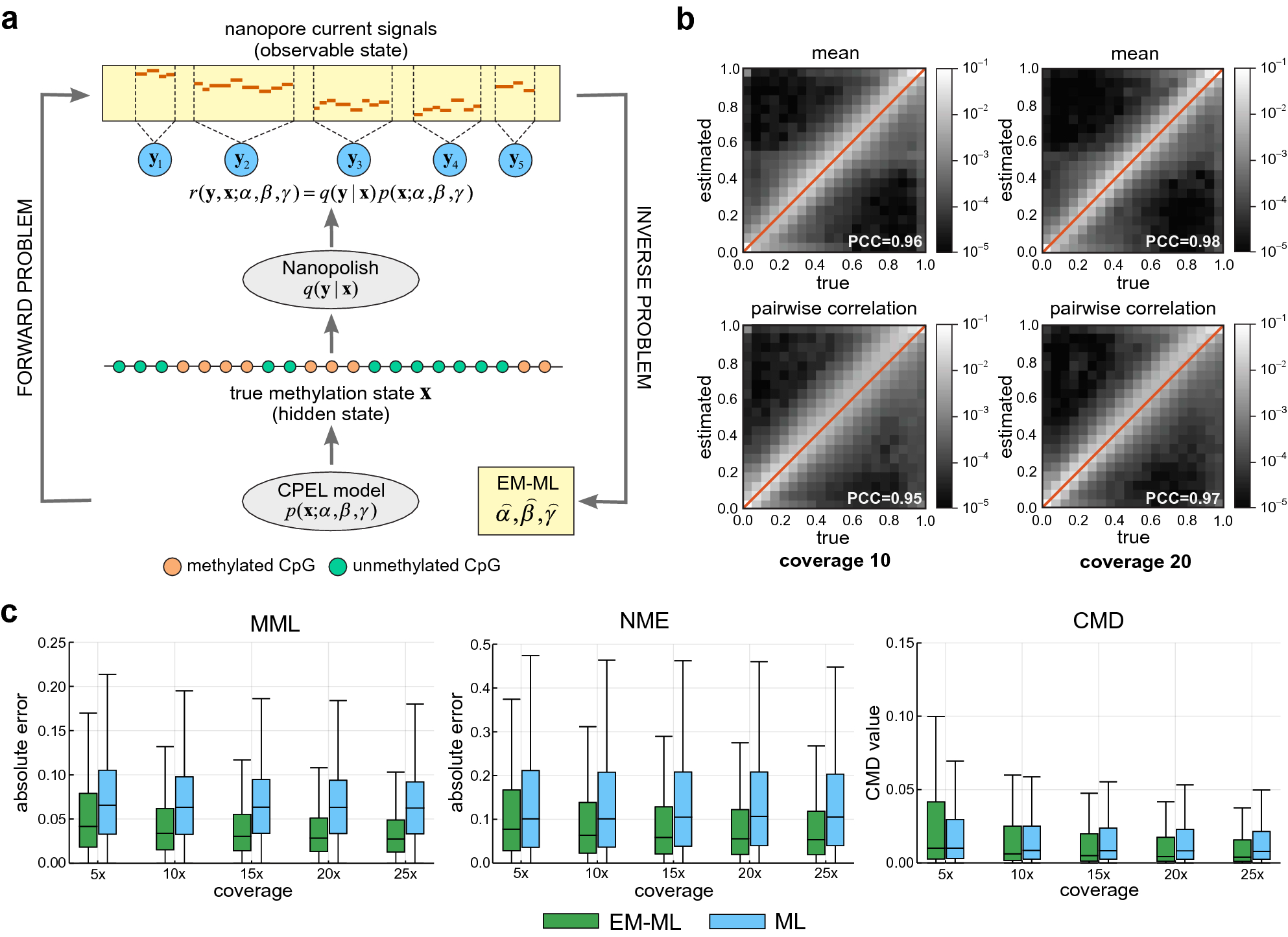
Estimating DNA methylation potential energy landscapes from nanopore sequencing data | Scientific Reports
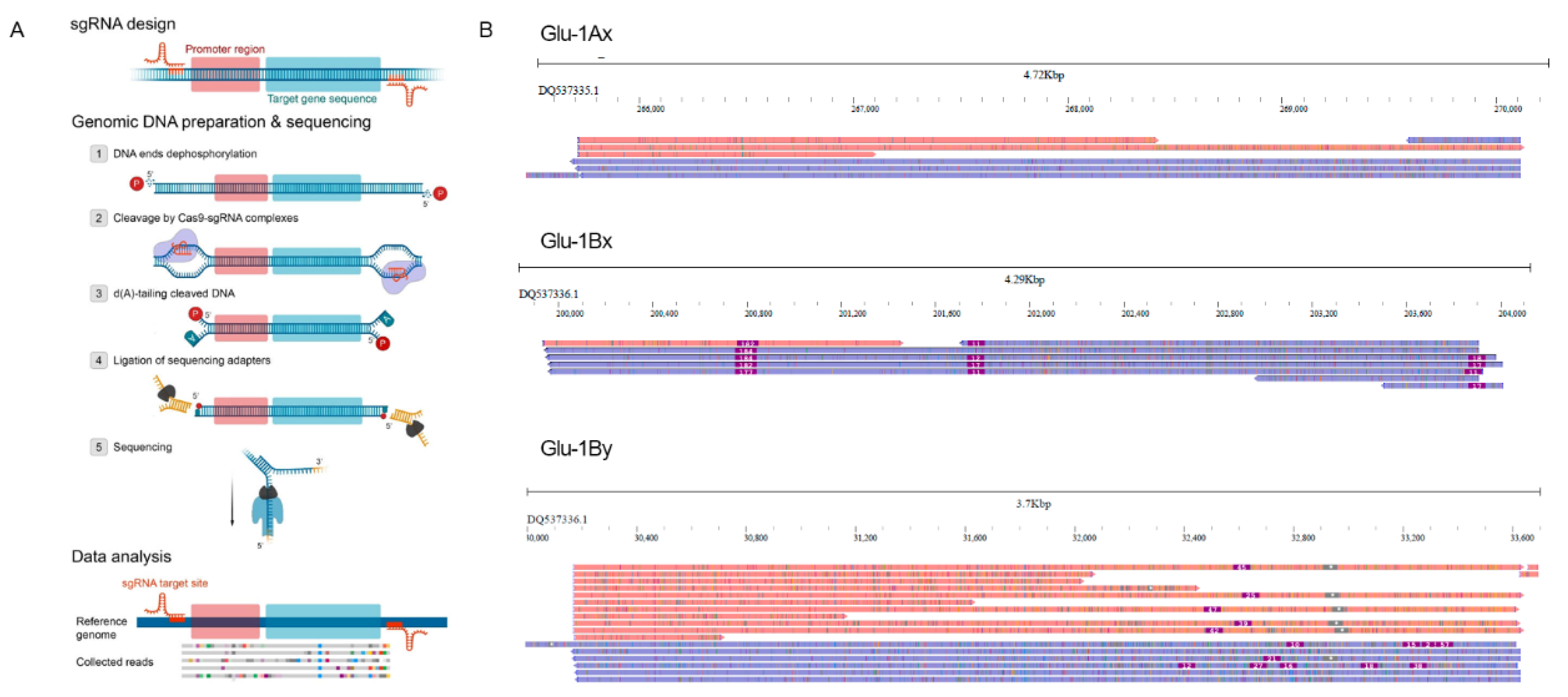
Plants | Free Full-Text | Searching for a Needle in a Haystack: Cas9-Targeted Nanopore Sequencing and DNA Methylation Profiling of Full-Length Glutenin Genes in a Big Cereal Genome